experiment = 1;
srcx = 30; srcy = 30;
baseDir = pwd;
baseDir = baseDir(1:end-3);
resultsDir = [baseDir 'results/'];
dataDir = [baseDir 'data/'];
rng('default');
if exist(resultsDir,'dir')==0
mkdir(resultsDir);
end
assert(exist('experiment','var')==1,'Need experiment variable');
assert(matlabpool('size')>0,'Need open matlabpool');
numWorkers = matlabpool('size');
eval(['spgLR_experiment' num2str(experiment)]);
v2struct(params);
rng(rand_seed);
switch freqIndex
case 75
dload([dataDir 'bgdata_freq75.mat']);
case 125
dload([dataDir 'bgdata_freq125.mat']);
end
nrecs = dims(1); nsrcs = dims(3);
m = nsrcs*nrecs; n = nsrcs*nrecs;
saveDir = resultsDir;
saveFile = [saveDir 'spgLR_experiment' num2str(experiment) '.mat'];
dload(saveFile);
spmd,
e = redistribute(e,getCodistributor(b));
bloc = getLocalPart(b); eloc = getLocalPart(e);
bloc(~eloc) = 0;
bsub = codistributed.build(bloc,getCodistributor(b));
bloc=[]; eloc = [];
end
src_idx = sub2ind([nsrcs,nsrcs],srcx,srcy);
pdims = [nrecs,nsrcs,nrecs,nsrcs];
P = oppPermute(pdims,[1 3 2 4]);
Xest = Lest * Rest';
spmd,Xest = redistribute(Xest,getCodistributor(b)); end
spmd
bloc = getLocalPart(b);
eloc = getLocalPart(e);
xloc = getLocalPart(Xest);
bNormTrain = norm(bloc(eloc))^2;
bNormTest = norm(bloc(~eloc))^2;
trainErr = norm(bloc(eloc)-xloc(eloc))^2;
testErr = norm(bloc(~eloc)-xloc(~eloc))^2;
recovery = [bNormTrain,bNormTest,trainErr,testErr];
xloc = []; bloc = []; eloc = [];
end
recovery = pSPOT.utils.global_sum(recovery); recovery = recovery{1};
trainErr = sqrt(recovery(3)/recovery(1));
testErr = sqrt(recovery(4)/recovery(2));
disp('-------SPGLR results-------');
disp(['Percentage missing receivers: ' num2str( subPercent*100 )]);
disp(['Rank: ' num2str(rank)]);
disp(['Total runtime: ' num2str( spgLRTime/3600 ) 'h']);
disp(['Number of workers: ' num2str( numWorkers ) ]);
disp(['Train (data fit) error: ' num2str(-20*log10(trainErr),3) 'dB']);
disp(['Test (recovery) error: ' num2str(-20*log10(testErr),3) 'dB']);
toStandardOrder = @(x) pSPOT.utils.distVec2distArray(P* pSPOT.utils.distVectorize(x),[nrecs^2,nsrcs^2]);
bsub = toStandardOrder(bsub);
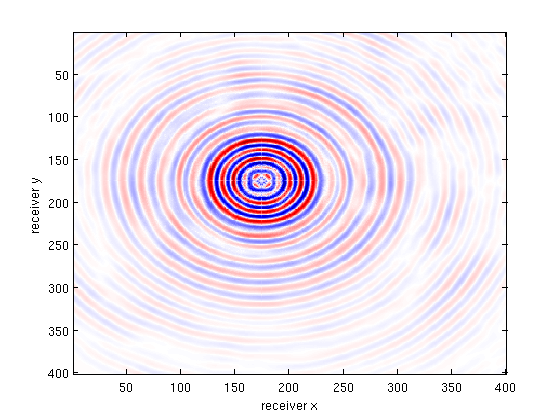
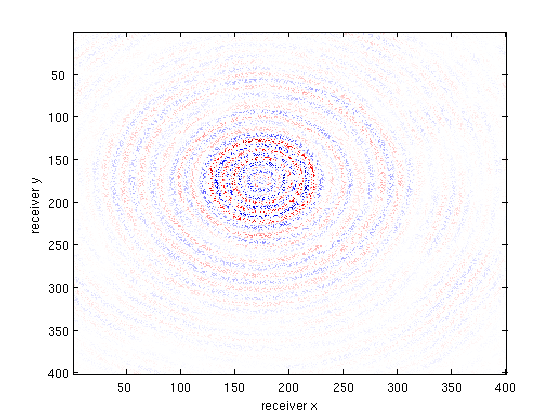
bsub_csg = reshape( gather(bsub(:,src_idx)),nrecs,nrecs);
b = toStandardOrder(b);
b_csg = reshape( gather(b(:,src_idx)) , nrecs, nrecs );
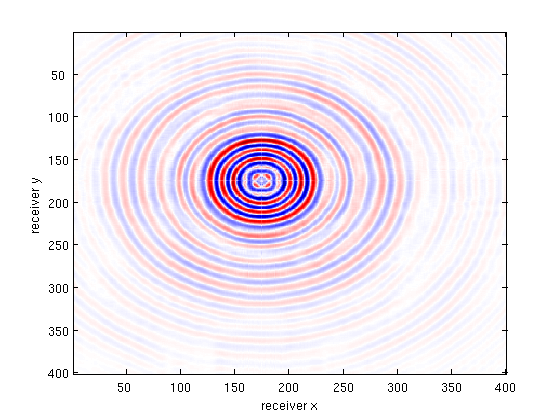
Xest = toStandardOrder(Xest);
X_csg = reshape( gather(Xest(:,src_idx)) , nrecs, nrecs );
figopts = struct;
figopts.xlabel = 'receiver x';
figopts.ylabel = 'receiver y';
figopts.caxis_ctr = true;
figopts.cmap = 'seismic_colormap';
close all;
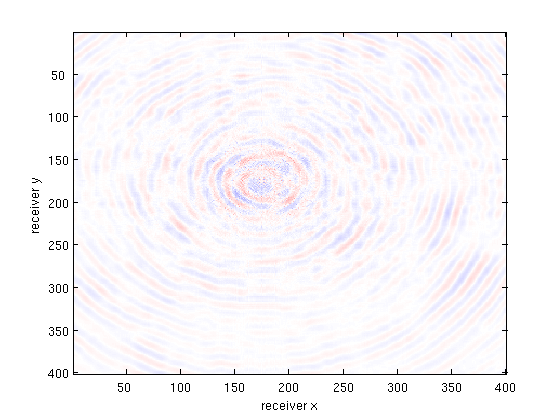
multi_imagesc(figopts,b_csg,bsub_csg,X_csg,b_csg-X_csg);
disp(['Slice SNR (data fit + recovery) : ' num2str(SNR(vec(b_csg),vec(X_csg)),3) 'dB']);
-------SPGLR results-------
Percentage missing receivers: 75
Rank: 500
Total runtime: 4.659h
Number of workers: 8
Train (data fit) error: 14.1dB
Test (recovery) error: 13.7dB
Slice SNR (data fit + recovery) : 13.8dB